Authors: Chris Lee1, Jonatan Lysen1, Jeff Roeder1, Julie Baggs1, Kiranmai Durvasula1, Travis Butts1
- Omega Bio-tek, Norcorss, GA 30071
Overview
Purpose
Development of a fully automated solution to extract cfDNA from up to 10 mL samples in 2.75 hours when integrated on Hamilton’s Microlab® STAR™ platform.
Methods
- cfDNA extracted using automated protocol with Mag-Bind® cfDNA Kit on a Hamilton Microlab STAR
- Yield and quality analysis using Agilent’s TapeStation® 2200
Results
- Efficient cfDNA purification achieved with automation protocol
- Automated protocol doubles cfDNA yield from manual protocol
- Average peak size was comaprable between automated and manual protocols
Introduction
The use of circulating, cell-free DNA (cfDNA) is rapidly evolving in th clinical landscape and is one of the fstest growing areas in recent years. It offers tremendous clinical potential as a screening method for tumor, cancer, fetal DNA studies, disease progression, treatment response, and more. Circulating DNA extraction is often challenging as they are found in low quantitities, and accurate methods are needed to isolated the less abundant cfDNA with higher sensitivity and specificity. Rapid, reliable, and high throughput methodologies are needed for widespread adoption solution to extract cfDNA from up to 10 mL samples in 2.75 hours when integrated on Hamilton’s Microlab STAR platform. The system is scalable and can extract sample volumes ranging from 1-10 mL without any hardware modification or dditional accessories on the Hamilton workstation.
Methods
cfDNA Extraction and Analysis
To evaluate the overall performance of the Mag-Bind cfDNA Kit, 10 mL of unspiked serum sample was used to isolate cfDNA using Omega Bio-tek’s Mag-Bind cfDNA Kit automated on a Hamilton Microlab STAR. NExt, to compare automated versus manual extraction protocols, cfDNA was extracted from 8 mL of sample using Omega Bio-tek’s automated protocol and from 4 mL of smaple using the manual extraction protocol. The automated extraction protocol follows the workflow presented in Figure 1. For the manual extraction, 4 mL of serum sample was used to extract cfDNA follwoing manufacturer’s protocols. Agilent’s TapeStation 2200 was used to evalutate the perfomance of each extration protocol.
Mag-Bind cfDNA Kit Extraction Workflow from 10 mL Input Automated on Hamilton Microlab STAR
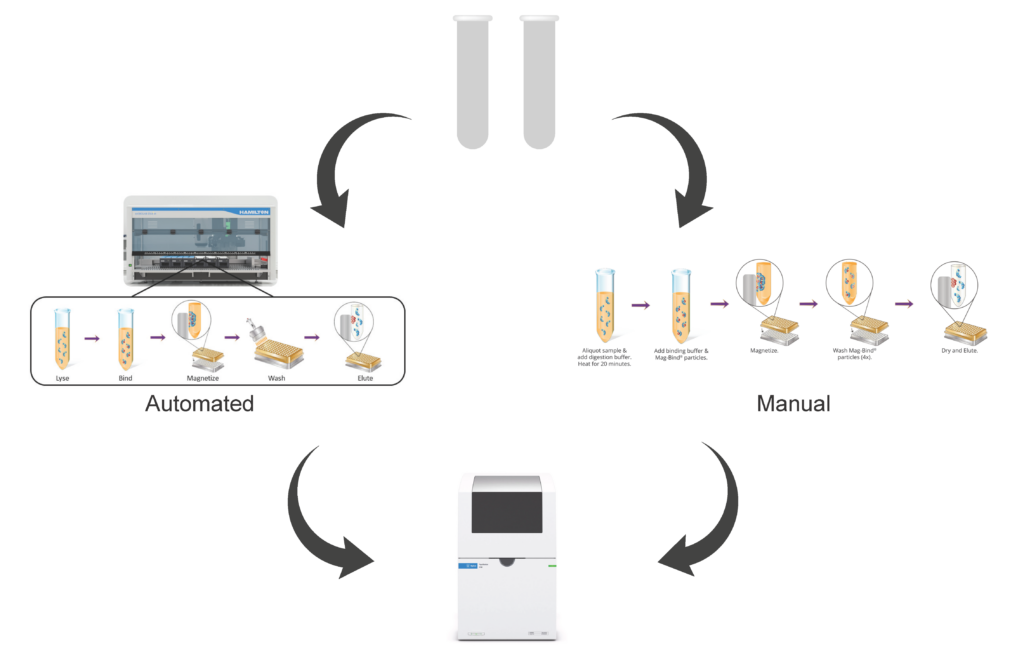
Figure 1. cfDNA was isolated from 10 mL of unspiked serum using Omega Bio-tek’s Mag-Bind cfDNA Kit automated on a Hamilton Microlab STAR.
Results
TapeStation Analysis of Purified cfDNA from 10 mL Plasma Samples
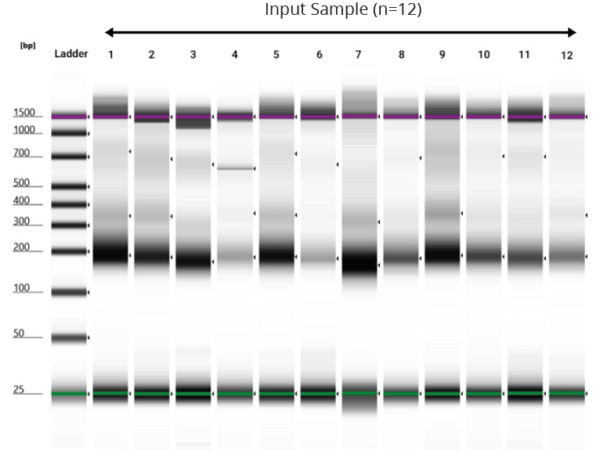
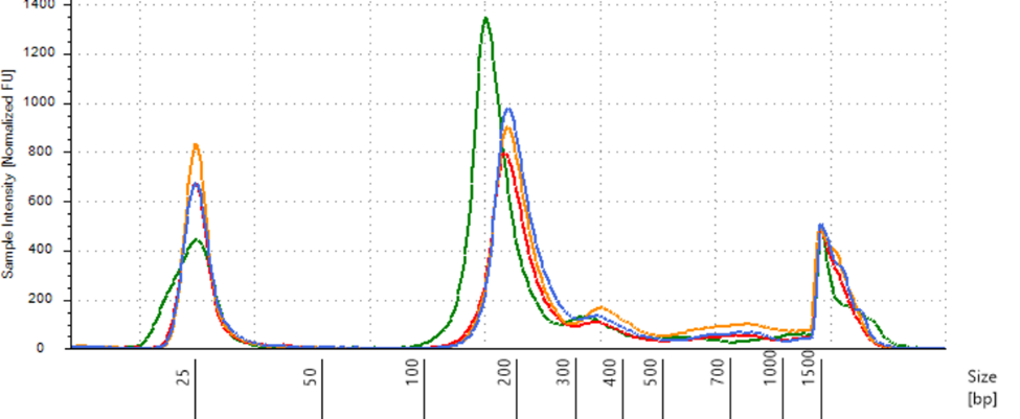
Figure 2. Gel image generated from TapeStation of cfDNA purified from 10 mL of unspiked plasma samples (n = 12) using Omega Bio-tek’s Mag-Bind cfDNA Kit automated on Hamilton Microlab STAR platform.Efficient purification of cfDNA was indicated by the characteristic peak at approximately 170 bp.
Table 1 Comparison of Yield Values from Manual vs Automated Extraction
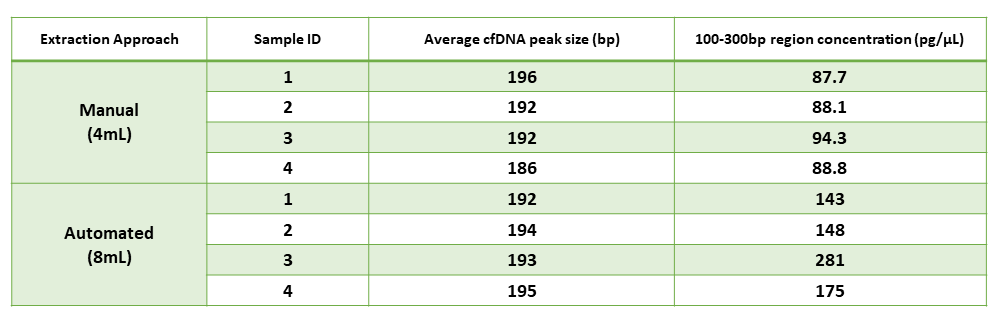
Regional Analysis using TapeStation software showed that cfDNA yield roughly doubled when doubling the sample volume from 4 mL in the manual extraction protocol to 8 mL in the automated protocol. The average cfDNA peak size was comparable between manual and automated extraction, showing ~192 bp versus ~194 bp, respectively.
TapeStation Analysis of Purified cfDNA from Manual vs Automated Extraction
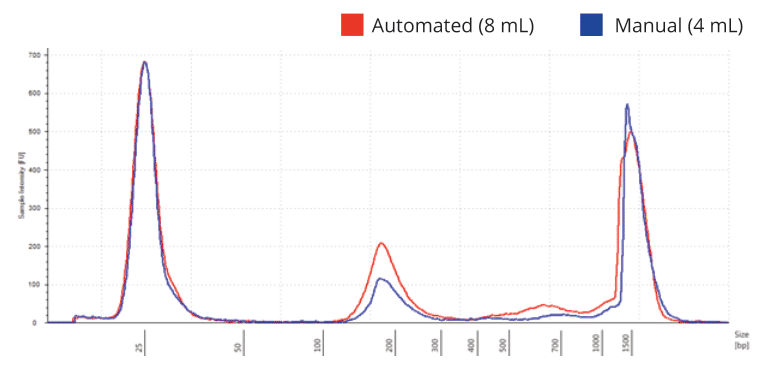
Figure 3. The overlay showed that the purified DNA contained more cfDNA and minimal genomic DNA contamination. Again, the average size of the cfDNA peaks was similar between the manual and automated potocols, with the automated protocol showing a higher intensity around the average peak of 194 bp.
Conclusions
From the findings presented, Omega Bio-tek’s high throughput workflow automated on the Hamilton Microlab STAR platform:
- allows flexible sample inputs and low elution volumes
- produces high quality cfDNA for use in various downstream applications
- addresses the need for a rapid, reliable, and high throughput method for extracting cfDNA
WP-0036
Originally presented at SLAS 2023
Download the poster here
