Victor Chen1, Kiranmai Durvasula2
1. Chroma ATE, Inc, Guishan Dist., Taoyuan City, Taiwan 333001
2. Omega Bio-tek, Inc, Norcross GA 30071
Introduction
Stool sample analysis offers a simple, non-invasive methodology for detection of multiple disease-causing microbes, biomarker detection of cancers of the digestive tract to analysis of gut microbiome and their impact on physiological health and disease. Molecular examination of the gut microbiome can provide valuable insight into the bacterial composition and can serve as an early screening tool to predicting treatment outcome of diseases of the gastrointestinal tract. However, stool is a complex sample source due to the presence of polyphenols, humic acid, lipids, and other PCR-inhibiting compounds. To realize its immense clinical potential, there is a need for extracting high quality DNA from stool free of PCR inhibitors in an automated, high throughput format. To address this need, Omega Bio-Tek has developed a versatile magnetic bead-based kit called Mag-Bind® Universal Pathogen 96 Kit for pathogen detection from various sample types including stool. In this application note, we provide the automated workflow for DNA extraction from stool specimens using Mag-Bind® Universal Pathogen 96 Kit on Chroma ATE’s MagXtract 3200. The product specifications of Chroma ATE’s MagXtract 3200 system are described in Table 1 below. The application note also discusses the performance of this automated workflow in terms of DNA yield, quality, and amplification potential using real-time PCR.
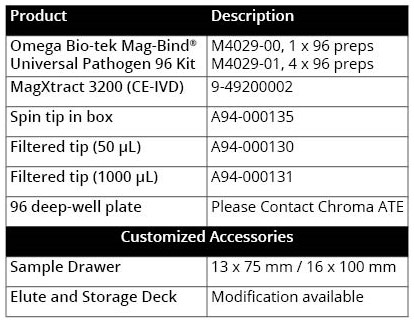
Table 1. Product specifications of Chroma ATE’s MagXtract 3200 system
Materials and Methods
25 mg of human, dog and cat stool samples were collected and diluted 10% w/v in double distilled water. The samples were mixed thoroughly. DNA from 250 μL of each of the stool samples was extracted using Omega Bio-tek’s Mag-Bind® Universal Pathogen 96 Kit automated on Chroma ATE’s MagXtract 3200 platform. The initial homogenization and lysis steps were performed manually offline. The rest of the protocol steps that include bind, wash, dry, elute as well as the qPCR set up were completely automated on MagXtract 3200. The system performs all the bead collection, transfer and dispense operations as necessary. The layout of the extraction workflow in a 96-well deep-well plate is as described in Figure 1.
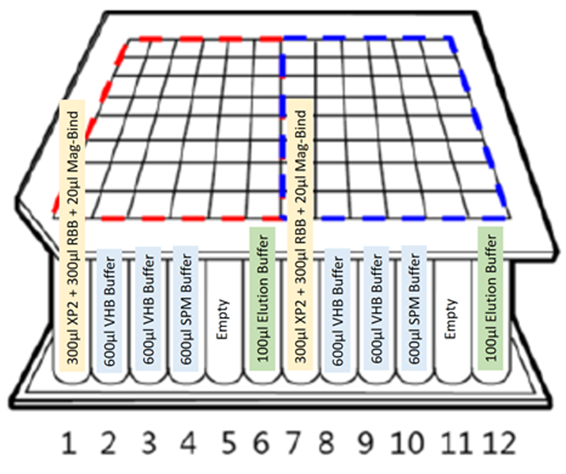
Figure 1. Extraction layout of the buffers in a 96-well deep-well plate on MagXtract 3200 platform.
DNA extracted from the three independent samples was eluted in 100 μL volume and quantified using Nanodrop ONE (ThermoFisher). A qPCR reaction was set up at three different input DNA template amounts of 5 ng, 0.5 ng and 0.05 ng to evaluate not only the downstream suitability of extracted DNA but also is amplification sensitivity. The qPCR utilized primers to detect the presence of Bifidobacterium in the extracted stool samples.
MagXtract 3200 Open System and Automation Solution
The MagXtract 3200 allows the authorized user to edit its protocols, including nucleic acid extraction and PCR preparation. The MagXtract 3200 is highly flexible and can easily fit into the workflows of most magnetic bead-based extraction kits and PCR reagent kits.
Open System
Following manufacturer’s protocols, the automated steps on MagXtract 3200 can be optimized to achieve good performance.
Automation Solution
Automation of the magnetic bead-based system reduces the number of manual steps required for extraction and PCR preparation.
GUI
The MagXtract 3200 has three operating modes: full run, preparation, and extraction.
MagXtract 3200 software provides protocol-based control to streamline workflows. The stepwise GUI and touchscreen control guide the user through the completion of the assay setup, from sample loading to the consumable placement.
Results and Discussion
The automated protocol time of 32 samples is 1 hour and 35 mins, including automatic distribution of primary samples to extraction deck and DNA extraction. The purified DNA yield from human, cat and dog stool samples determined using Nanodrop ONE is as shown in Table 2. The A260/A280 ratios for cat and dog stool samples were 1.9 and 1.3 for human stool sample suggesting high-quality of extracted DNA (Table 2).
The suitability of the purified DNA for downstream applications were determined by running a qPCR reaction. Figure 2 shows the average Ct values obtained on 5 ng (1X), 0.5 ng (10X) and 0.05 ng (100X) of the purified DNA using Bifidobacterium specific primers. The amplification curves in Figure 2 indicate good qPCR efficiency. The ΔCt between the DNA dilutions (10X and 1X, 100X and 10X) for all the stool samples is ~3.3, illustrating the DNA is free of inhibitors and is of high-quality.
Table 2. Yield and purity of extracted genomic DNA from independent stool samples.
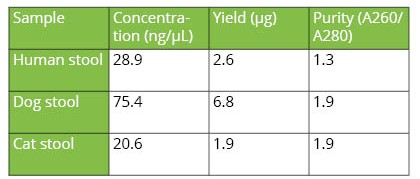
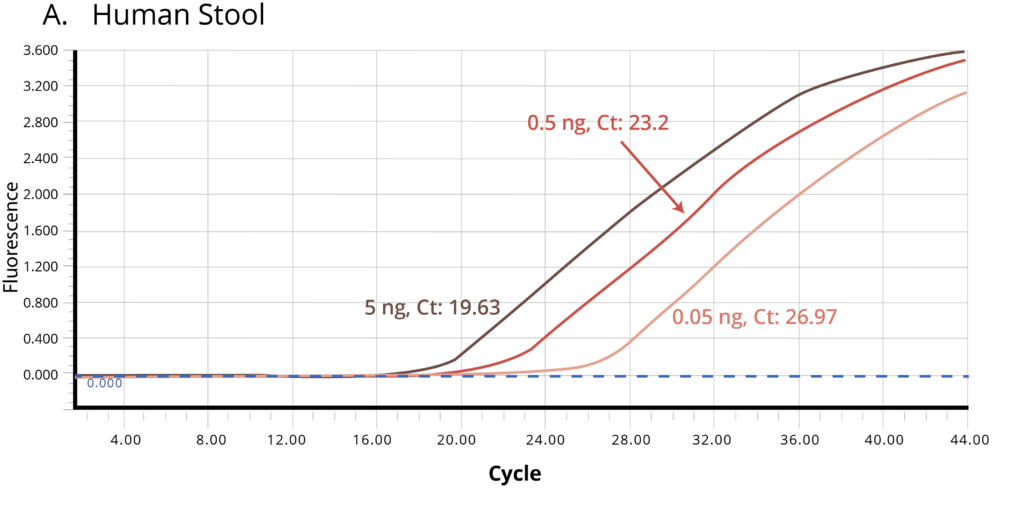
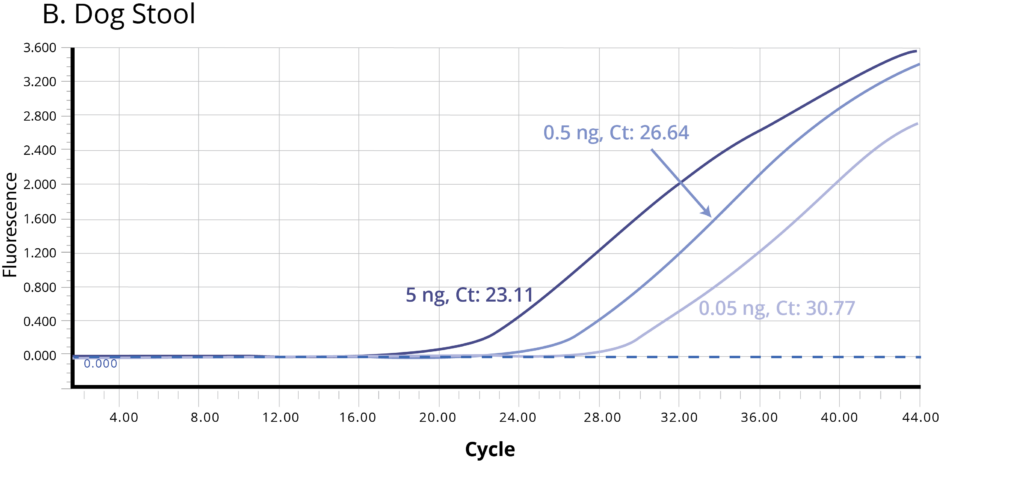
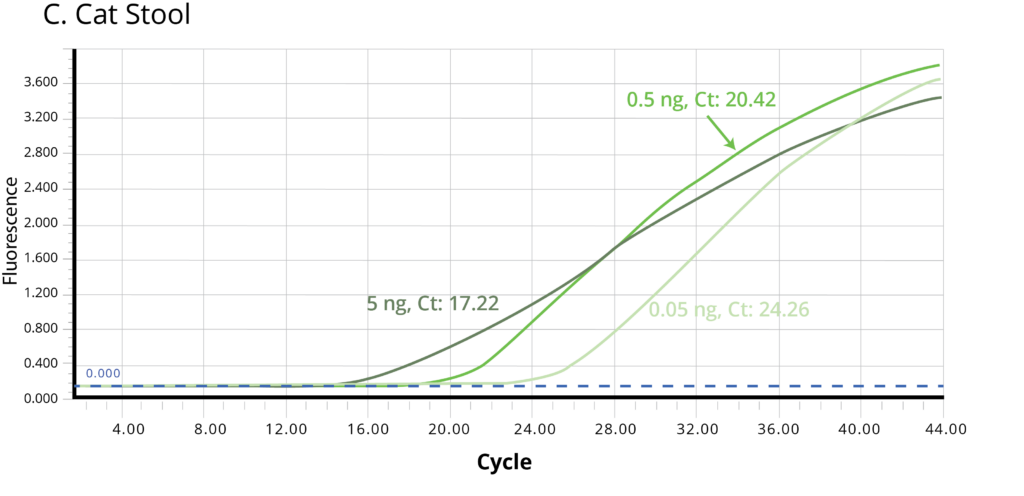
Figure 2. Stool samples from three independent donors, A. Human, B. Dog, and C. Cat, respectively, were analyzed using qPCR to detect Bifidobacterium. Each of the gDNA templates in the qPCR reactions were normalized to 0.05, 0.5, and 5.0 ng/reaction.
Conclusions
Omega Bio-tek’s Mag-Bind® Universal Pathogen 96 Kit automated on the MagXtract 3200 offers a streamlined approach for isolating genomic DNA from stool samples and also setting up qPCR from extracted DNA. This workflow can potentially save 80% of the hands-on time required. Purified DNA is inhibitor-free and is suitable for various downstream applications such as PCR, restriction digestion, and NGS.
Product Information
WP-0039


