Research publication review and discussion: “Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring”, Nature Medicine.
 Circulating, cell-free DNA (cfDNA) holds great clinical significance for non-invasive disease detection, diagnosis, and monitoring for tumor, cancer, or fetal DNA studies. Circulating DNA extraction is often challenging as they are found in low quantities, and accurate, consistent methods are needed to isolate the less abundant cfDNA with higher sensitivity. Cancer and oncology researchers worldwide use cfDNA analysis to advance oncological research and make informed treatment decisions. In this context, Omega Bio-tek has developed cfDNA extraction kits for both manual processing using silica spin columns and for automated, high-throughput workflows using magnetic beads. In this blog post, we will see how Omega’s Mag-Bind® cfDNA Kit played a vital role as the superior cfDNA extraction method used in a recent publication in Nature Medicine.
Circulating, cell-free DNA (cfDNA) holds great clinical significance for non-invasive disease detection, diagnosis, and monitoring for tumor, cancer, or fetal DNA studies. Circulating DNA extraction is often challenging as they are found in low quantities, and accurate, consistent methods are needed to isolate the less abundant cfDNA with higher sensitivity. Cancer and oncology researchers worldwide use cfDNA analysis to advance oncological research and make informed treatment decisions. In this context, Omega Bio-tek has developed cfDNA extraction kits for both manual processing using silica spin columns and for automated, high-throughput workflows using magnetic beads. In this blog post, we will see how Omega’s Mag-Bind® cfDNA Kit played a vital role as the superior cfDNA extraction method used in a recent publication in Nature Medicine.
In July 2020, a group of researchers from prestigious institutions such as the Weil Cornell Medical College, New York; the Fred Hutchinson Cancer Research Center, Seattle; and the New York Genome Center published an article in Nature Medicine titled, “Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring.” Asaf Zviran et al. wrote about how low cfDNA abundance is a barrier and presented an orthogonal framework for cfDNA cancer monitoring via genome-wide mutational integration, enabling ultra-sensitive detection, overcoming the limitation of cfDNA abundance, and empowering treatment optimization in low-disease-burden oncology care.1
In this post, we will look at a general overview of this 37-page behemoth publication and note how Omega Bio-tek’s Mag-Bind® cfDNA Kit provided better performance and ease of use compared to other tested methods1 helping researchers with the challenges of low cfDNA abundance and using whole genome sequencing (WGS) for tumor burden tracking and post-operative disease detection.
Hypothesis
Zviran et al. postulated that limited input material may be the fundamental barrier to detection sensitivity and hindering deep targeted approaches. The limited input material is the number of cfDNA fragments as measured by genomic equivalents (GEs). Genomic equivalents are the amount of DNA necessary to be present in a purified sample to guarantee that all genes (in this case, genes associated with cancer detection) will be present. Specifically, in the setting of early cancer or minimum residual disease (MRD) detection, cfDNA abundance might be limited to deep targeted sequencing, as it imposes a ceiling on effective depth of sequencing.1 Targeted sequencing uses deep sequencing to find known and new variants within the region of interest.
To test this hypothesis, they gleaned insight from a recent landmark study of early-stage cancer detection. It found mutation detection was limited mainly to 10−3 variant allele fraction (or VAF, used to determine whether a variant comes from somatic or inherited) in cfDNA for all cancer stages. This VAF is despite the ~40,000× coverage depth of sequencing and the application of advanced molecular error suppression. This sensitivity limitation manifested in a stage-dependent decrease in the proportion of patients with detectable ctDNA (circulating tumor DNA, a specific type of cfDNA).1

Problem
A hindrance to testing this hypothesis is in the setting of low-burden disease detection—where tumor fraction (TF) is low—limited input material likely constitutes a significant barrier to the practical application of deep targeted sequencing. Analysis of TF is critical for the detection of MRD, which guides clinical intervention. However, cfDNA TF is significantly lower in the MRD context as it is correlated with disease burden.1 Additionally, many patients with radiographically evident disease do not show detectable ctDNA by deep targeted sequencing. 1
Possible Solutions
Since researchers cannot overcome the limited cfDNA input through sample processing improvement, researchers turned their focus on library and analysis design to maximize the use of cfDNA fragments. The rest of the publication discusses different arenas of library and analysis design optimization. One aspect explored was that the breadth of sequencing supplants depth of sequencing to overcome the low number of DNA in plasma samples. Another was the development and validation of ultra-sensitive ctDNA detection via genome-wide mutational integration. Next, expanding the genome-wide mutational integration approach to copy number alterations was discussed. Additionally, researchers examined the application of the genome-wide mutational integration approach to clinical plasma samples. Finally, researchers looked at how genome-wide mutational integration enables the detection of post-operative residual disease.1
Discussion
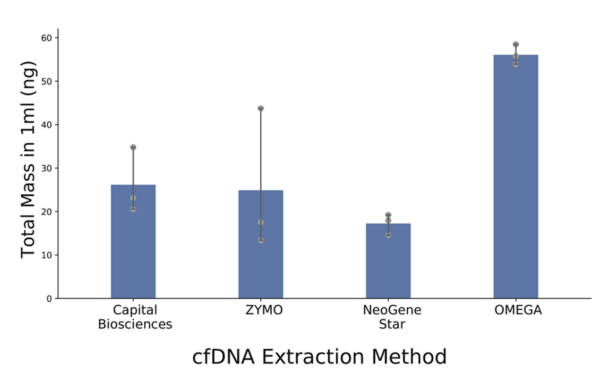
Notably, Omega Bio-tek’s Mag-Bind® cfDNA Kit was the superior product for cfDNA extraction method over the kits from three other tested companies. The Mag-Bind® cfDNA Kit displayed overall somewhat better performance and ease of use compared to other tested methods. Therefore, the research group elected to perform Omega Bio-tek extraction for patient and control plasma samples. 1
About Omega Bio-tek
Omega Bio-tek, founded in 1998, is an ISO 9001:2015 industry-leading manufacturer of DNA/RNA purification kits for clinical, biotechnology, and genomics research. With a diversified product portfolio for low-throughout to high-throughput purification, Omega Bio-tek kits purify high-quality nucleic acids from a wide variety of samples. Omega Bio-tek’s products and services are used in over 1000 universities and research institutes and over 100 different countries worldwide.
Omega Bio-tek is dedicated to helping high-throughput researchers transform ideas into revolutionary life science advancements by continuing to provide an expanding product portfolio and a wide of automation solutions.
For more information on the Mag-Bind® cfDNA Kit used in this study, visit
https://www.omegabiotek.com/product/mag-bind-cfdna-kit/

References
- Zviran et al. 2020. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nature Medicine; 1114: 1114-1124.
