Mag-Bind® SeqDTR
$0.00 – $11,332.20
Removal of unincorporated terminators from sequencing reactions using magnetic beads
- No protocol change against major competitor
- Read lengths averaging over 800 bps (Min Phred 20)
- Manual or adjustable to automated liquid handlers
- Significant cost savings compared to sephadex-based clean ups
- 96- or 384-well formats
Mag-Bind® SeqDTR is designed to effectively and reliably remove unincorporated terminators from sequencing reactions. The system combines Omega Bio-tek’s proprietary chemistries with the reversible nucleic acid-binding properties of magnetic beads to eliminate excess nucleotides, primers, and small, non-targeted amplification products such as primer dimers. Sequencing products are mixed with the Mag-Bind SeqDTR magnetic particles, which selectively bind DNA. Two rapid wash steps eliminate trace contaminants such as nucleotides and primers to reduce background signal and, therefore, achieve higher QV scores.
The high sensitivity of Mag-Bind SeqDTR’s binding ability allows for decreased concentrations of BigDye® chemistry to be used and longer continuous read lengths to be achieved. Mag-Bind SeqDTR can be processed in 96- and 384-well formats and is compatible with many liquid handling instruments, including Hamilton Microlab® STAR™ & STARlet™, Beckman Coulter Biomek® FX & NX, and Tecan Evo instruments. Up to 4 plates can be run in a 96-well format in less than 25 minutes.
For Research Use Only. Not for use in diagnostic procedures.
| FEATURES | SPECIFICATIONS |
|---|---|
| Downstream application | Sanger Sequencing, Cloning, In Vitro Transcription, Nucleic Acid Labeling, PCR, Real-Time Quantitative PCR (qPCR), Sequencing, Southern Blotting |
| Elution volume | 40 µL of Elution Buffer for 96-well plate protocol. 15-20 µL of Elution Buffer for 384-well plate protocol. |
| Starting material | ABI Big Dye Chemistry Cycle Sequencing Reactions |
| Starting amount | 5 – 20 µL |
| DNA recovery | Sequencing reads start at 40 nt |
| Processing mode | Automated; Manual |
| Throughput | 96-384 samples per run |
| DNA binding technology | Magnetic beads |
| Binding capacity | Scalable |
| Storage | 2°C – 8°C |
Superior Signal Strength
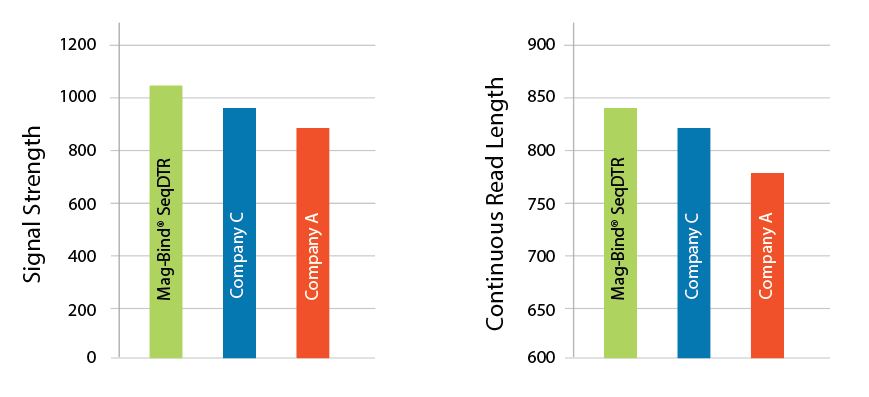
Figure 1. Purified 1.8 kb PCR fragments were sequenced from each company using the manufacturer’s recommended protocols. The median of 16 samples per company are used in the representations above. A 5 µL sequencing reaction was performed using a 1/32 dilution of Applied Biosystems BigDye Terminator v3.1 chemistry. DNA was analyzed on an Applied Biosystems 3730XL.
Cost Comparison
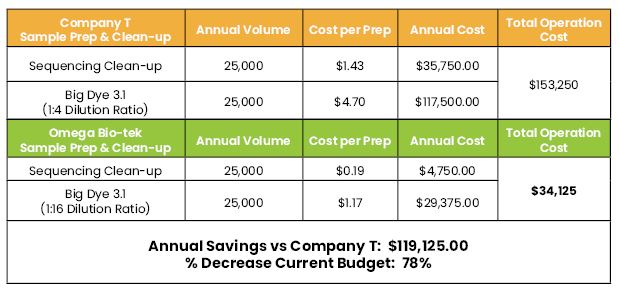

Table 1. Less BigDye is needed when using Mag-Bind SeqDTR for sample prep and clean up. Therefore reduce cost in BigDye usage. Mag-Bind SeqDTR cost per prep is significantly lower than the competitor.
Product Information
Safety Data Sheets
| Components | Hazard Standards | Languages | Link | hf:tax:dlp_document_language | hf:tax:dlp_document_hazard-standard |
|---|---|---|---|---|---|
| Mag-Bind SeqDTR | GHS | English | english | ghs | |
| Mag-Bind SeqDTR | GHS | Spanish | spanish | ghs | |
| Mag-Bind SeqDTR | REACH | English | english | reach | |
| Mag-Bind SeqDTR | REACH | Danish | danish | reach | |
| Mag-Bind SeqDTR | REACH | Finnish | finnish | reach | |
| Mag-Bind SeqDTR | REACH | French | french | reach | |
| Mag-Bind SeqDTR | REACH | German | german | reach | |
| Mag-Bind SeqDTR | REACH | Italian | italian | reach | |
| Mag-Bind SeqDTR | REACH | Norwegian | norwegian | reach | |
| Mag-Bind SeqDTR | REACH | Spanish | spanish | reach | |
| Mag-Bind SeqDTR | REACH | Swedish | swedish | reach | |
| Mag-Bind SeqDTR | WHMS | English | english | whms | |
| Mag-Bind SeqDTR | WHMS | French | french | whms |