Chris Lee¹, Sara Amirahmadi¹, Tesfaye Kemsi¹, Jeff Roeder¹, and Travis Butts¹
- Omega Bio-tek, Norcross, GA 30071
Introduction
Cell-free DNA (cfDNA) has emerged as a revolutionary tool in the realm of molecular diagnostics, offering a non-invasive window into the genetic information of various bodily tissues. The development of cfDNA technology is a large step forward in detecting and profiling circulating biomarkers to derive clinically-relevant information. However, researchers and clinicians alike face challenges when attempting to extract high-quality cfDNA in a streamlined manner due to its low concentrations and short, fragmented nature. For efficient implementation of applications downstream, a high throughput method for purifying high-quality cfDNA is critical. To address these obstacles, Omega Bio-tek has developed a high throughput automated workflow configuration to extract cfDNA from ninety-six, 4 mL plasma samples in approximately 3 hours. The Mag-Bind® NAP STAR is an Assay-Ready Workstation specifically configured to run Omega Bio-tek’s Mag-Bind® DNA purification Kits using Hamilton’s Microlab® STAR™. Here, we elucidate the methods of the Mag-Bind® NAP STAR using Omega Bio-tek’s Mag-Bind® cfDNA Kit (M3298) and compare its performance to that of extractions performed manually using the same Kit.
Learn more about the Mag-Bind® cfDNA Kit.
Materials and Methods
Omega Bio-tek’s Mag-Bind® NAP STAR automates cfDNA purification from 4 mL of plasma on the Hamilton Microlab® STAR™. This protocol processes 1-4 sample plates and is compatible with common cfDNA collection tubes, such as Streck Cell-Free DNA BCT® or Roche Cell-Free DNA Collection Tubes. The workflow was automated using a Hamilton Microlab® STAR™; the components equipped, their purpose, and positions on the deck are mapped in Figure 1.
Three 4 mL aliquots were sampled from three lots of human-derived plasma for extractions using 4×24-well plates. Circulating cell-free DNA was extracted manually using Omega Bio-tek’s Mag-Bind® cfDNA Kit following manufacturer’s instructions. Following this, cfDNA was extracted using an automated protocol for 4×24-well plates on the Hamilton Microlab® STAR™ using the same Omega Bio-tek Kit.
cfDNA purified using both manual and automated protocols was analyzed on Agilent’s TapeStation® 2200 for yield and quality. The samples extracted with the protocol utilizing 4×24-well plates were also subjected to qPCR for comparison to manual extraction methods and their efficacy in downstream applications.
Hamilton Microlab® STAR™ Deck Layout
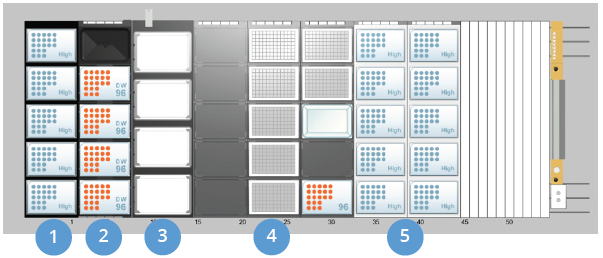
| Component | Purpose |
|---|---|
| 1. Tip Isolator Carrier | For tips reused for tip mixing and liquid waste removal steps |
| 2. MFX Carrier with 4x tall labware locators & liquid waste module for MPH | Magnet positions (Alpaqua Magnum FLX24) & liquid waste module |
| 3. HHS Baseplate with 4x HHS units (3 mm orbital with flat bottom adapters) | Heater/Shakers for bead resuspension and incubation steps |
| 4. 3x DWP Stands | For processing plate positions, reagent reservoirs, and elution plate |
| 5. 2x Standard Tip Carriers | Tips for reagent dispenses |
Figure 1. Hamilton Microlab® STAR™ deck layout for extraction of cfDNA from 4 mL plasma samples.
Results and Discussion
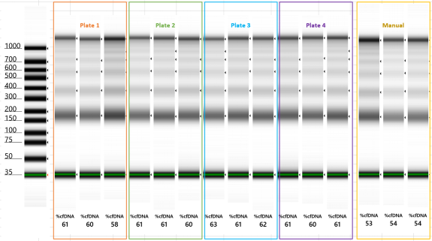
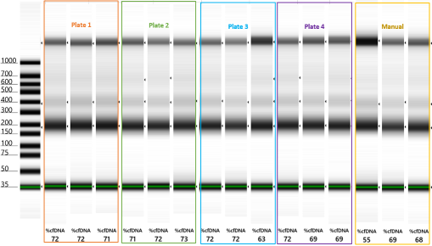
cfDNA purified using both manual and automated extraction protocols were analyzed on Agilent’s TapeStation® 2200 to derive information about the cfDNA’s size and integrity (Figures 2-4). %cfDNA was determined by the TapeStation® 2200 analysis software. Across all three lots of plasma, the %cfDNA extracted using the Mag-Bind® NAP STAR was greater than or equal to %cfDNA extracted manually. Figure 2 shows TapeStation® analysis of cfDNA extracted from plasma Lot A; these extractions produced a well-defined band at approximately 170 bp, the typical peak of cfDNA. Figures 3 and 4 support the same conclusion, as each TapeStation® image illustrates proper migration with well-defined bands around 170 bp and additional multimers above.
TapeStation Analysis of cfDNA Purified from Plasma Lot A

TapeStation Analysis of cfDNA Purified from Plasma Lot B
TapeStation Analysis of cfDNA Purified from Plasma Lot C
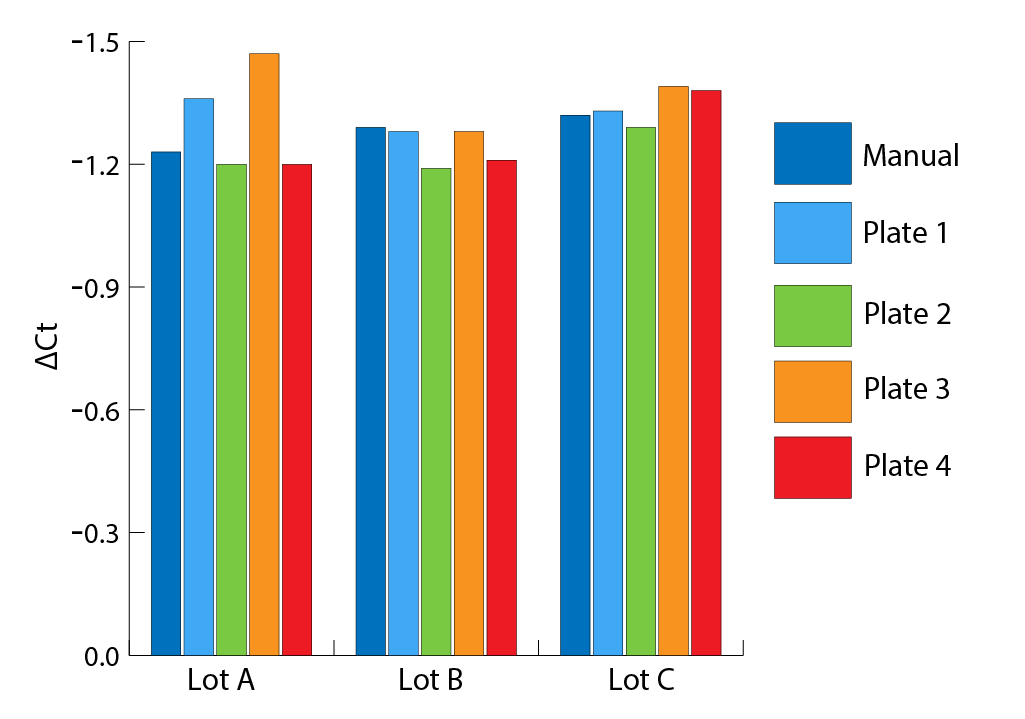
qPCR analysis was performed on the extracted cfDNA to determine its suitability for use downstream. Figure 5 presents the ΔCt values obtained from this analysis. Because of the 2 μL and 6 μL template input volumes, the ΔCt’s should theoretically be ~1.5. It can be seen in Figure 5 that extractions performed using Omega Bio-tek’s automated protocol exhibit ΔCt’s near the expected value of 1.5, indicating little to no inhibition; these results are also comparable to those obtained with manual extraction.
ΔCt Values from qPCR Analysis Illustrate Suitability for Downstream Applications
Conclusions
Omega Bio-tek’s Mag-Bind® NAP STAR offers an automated, high throughput purification solution for cfDNA from 4 mL plasma samples. This automation configuration produces high-quality cfDNA which is comparable to the cfDNA extracted using the manual protocol. Using this workflow, ninety-six 4 mL plasma samples can be processed in approximately 3 hours, starting with samples already transferred to the 24-well plate. The high yield and quality of the purified cfDNA support its suitability for use in downstream applications, such as qPCR.
WP-0058
Related Products
-
Circulating DNA
Mag-Bind® cfDNA Kit
$0.00 – $2,675.20Price range: $0.00 through $2,675.20 Select options This product has multiple variants. The options may be chosen on the product page

