Kiranmai Durvasula¹, Jessie Chen¹, Julie Baggs¹, Travis Butts¹
¹Omega Bio-tek, Inc., Norcross, GA 30071
Introduction
Formalin-fixed paraffin-embedded (FFPE) tissue samples are invaluable biospecimens that are increasingly being used in molecular level analyses and gene expression studies aiding the fields of cancer genomics, pathological diagnosis, biomarker discovery, immunotherapy research and basic research. Obtaining high quality and high yielding DNA and RNA is challenging since nucleic acids in FFPE tissue samples are often fragmented and prone to chemical modifications. To answer this need, Omega Bio-tek has developed an innovative nucleic acid extraction methodology (Mag-Bind® FFPE DNA/RNA 96 Kit – M6955) that not only partially reverses the formaldehyde-induced crosslinking but also extracts DNA and RNA in separate eluates from the same sample source without the need for sample splitting. Mag-Bind® FFPE DNA/RNA 96 Kit follows a magnetic bead-based approach and is automation-friendly. The protocol uses non-toxic mineral oil in combination with heat for efficient deparaffinization of the FFPE sample and differentially purifies DNA and DNA-free RNA allowing for a more comprehensive molecular analysis from a precious sample source. Here, we discuss the efficacy of using the Omega kit for extraction of both DNA and RNA from various FFPE tumor samples and compare its performance with other manufacturer’s comparable kits in terms of yield and quality, amplification potential and sensitivity of detection using real-time PCR. The quality of extracted DNA and RNA from the three different kits was evaluated in terms of their sequencing potential by generating NGS data with Illumina’s TruSight™ Tumor 170 panel covering a wide range of tumor genes and variant types.
Learn more about the Mag-Bind® FFPE DNA/RNA Kit.
Materials and Methods
DNA and RNA extraction
FFPE sections from three different tumor samples (breast, lung and colorectal tissues) were sourced from ProteoGenex (Inglewood, CA). DNA and RNA were extracted from single 10 µm sequential sections of each of these cancerous samples using Omega Bio-tek’s Mag-Bind® FFPE DNA/RNA 96 Kit and comparable kits from Company T and Company Q following manufacturer’s recommended protocols. Omega Bio-tek’s and Company T’s extraction protocol was magnetic-bead based whereas the protocol was silica column-based for Company Q. Non-toxic mineral oil was employed for the deparaffinization of the FFPE tissue sample in the Omega Bio-tek protocol whereas xylene was used for the other two kits. Extractions were carried out in triplicate from the same FFPE tissue sample in a single workflow for all the three kits. Purified DNA and RNA was eluted in 100 µL following protocols from Omega Bio-tek and Company T and in 50 µL following Company Q’s protocol.
Quantification, Quality, and Functionality Assessment
The purified nucleic acids were quantified using Thermo Scientific’s NanoDrop™ 2000c system and Promega’s QuantiFluor® dsDNA and RNA systems for specific quantitation of DNA and RNA without any interference from each other. Nucleic acid purity was analyzed by focusing on A260/A230 and A260/A280 absorbance ratios. The downstream functionality of the extracted nucleic acids was assessed by performing real-time PCR on 10-fold dilution of samples. Purified RNA was first transcribed into complementary DNA (cDNA) through reverse transcription and amplified using exon spanning primers specific for cDNA for accurate RNA analysis. A 20 µL qPCR reaction was set up using Agilent’s Brilliant III 2X SYBR® as the master mix and 2 µL of DNA or cDNA at appropriate dilutions as template and amplified with specific primers following a standard protocol on the ABI 7900.
Next-Generation Sequencing
Illumina’s TruSight™ Tumor 170 panel allows for comprehensive analysis of extracted DNA detecting small variants, amplifications, fusions etc covering 170 genes associated with solid tumors. Depending on the sample and extraction methodology, DNA input into this assay ranged from 35 ng to 120 ng and was sequenced on a HiSeq™ X Sequencing System.
Formalin-fixation and paraffin-embedding process introduces cross-linking and fragmentation through chemical modification in the nucleic acids, thereby compromising their quality. To evaluate the success of DNA and RNA extracted from an FFPE sample in a sequencing scenario, scientists from Illumina have put forth metrics like the ΔCq value for DNA and DV200 for RNA. The ΔCq value is calculated by comparing the amplification potential of an FFPE sample relative to that of an equivalent non-FFPE sample and the DV200 value represents percentage of RNA fragments that are greater than 200 nucleotides in length. These metrics serve as guidelines for success estimation in a sequencing workflow. Illumina estimates the success rate to be close to 100% with a ΔCq <5 and a DV200 ≥ 20 [1]. The fragment sizes and quality of the extracted RNA was analyzed on Agilent’s TapeStation® 2200 by running an RNA tape to derive a DV200 value. DNA extracted from fresh frozen colorectal tumor sample was included as a control for FFPE colorectal tumor tissue sample in this sequencing assay as a representative for ΔCq calculation. It was estimated as the difference in Ct between the FFPE and the non-FFPE sample by using a 2 µL template of a 10-fold diluted DNA normalized to 25 ng/µL in a 20 µL qPCR reaction.
Results and Discussion
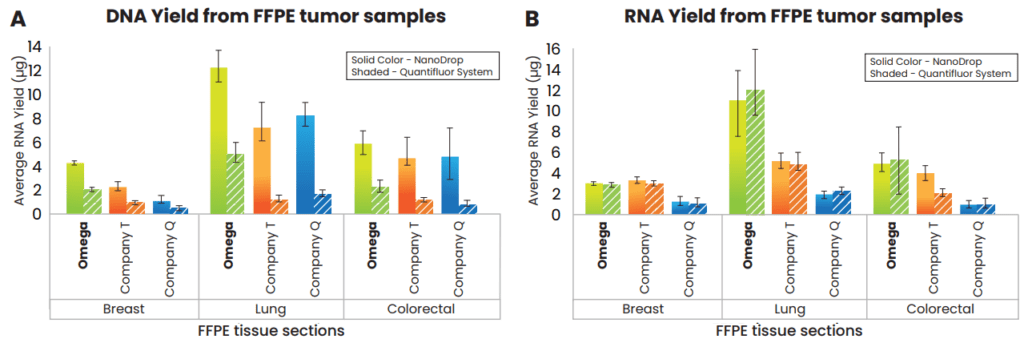
The average DNA and RNA yields quantified on NanoDrop™ 2000c system as well as Promega’s QuantiFluor Systems from FFPE sections of breast, lung and colorectal tumor tissue samples using the different manufacturer’s kits are as shown in Figure 1. As the fluorescent based dyes are sensitive enough to distinguish between DNA and RNA, it is expected to find the yields lower or comparable to the spectrophotometric measurements made using NanoDrop (Figure 1). A Tukey’s post-hoc analysis suggests that the average DNA yield following Omega Bio-tek’s protocol for FFPE lung and breast tumor samples was significantly higher compared to the average DNA yield obtained following protocols from Company T and Company Q (p<0.05). The average DNA yield was comparable for the FFPE colorectal cancer sample for all the three extraction methodologies tested. As for the average RNA yield, Omega Bio-tek’s was significantly higher for FFPE lung tumor sample compared to Company T and was significantly higher than Company Q’s for all the FFPE tumor samples tested (p<0.05; Tukey’s post-hoc analysis). The average RNA yield for FFPE breast and colorectal tumor tissue samples was comparable using kits from Omega Bio-tek and Company T.
The absorbance ratios of the nucleic acids (A260/A280 and A260/A230) are as shown in Figure 2 and were used to assess the DNA and RNA purity post-extraction using the three different kits. The average A260/A280 for DNA and RNA following Omega Bio-tek’s protocol was close to the theoretical 1.8 and 2.0 indicative of pure DNA and RNA (DNA – 1.84, 1.82, 1.86 and RNA – 1.93, 1.98, 2.0 for breast, lung and colorectal tumor tissue samples respectively) (Figure 2). These ratios also indicate efficient separation of DNA and RNA into separate eluates during the extraction protocol. For all the FFPE tumor tissue samples, the A260/A280 ratio for DNA and RNA extracted using kits from Company T and Company Q was close to 2.0 suggesting possible RNA contamination in the DNA eluate. A secondary purity measurement of A260/A230 was made and the average for the purified DNA and RNA ranged between 1.33 and 1.72 for Omega and was less than 0.64 for Company T. As for Company Q, the ratio of A260/A230 was >2.0 for DNA and <0.60 for RNA. These values suggest that the nucleic acids extracted using the Omega Bio-tek kit were of superior quality than those extracted using the other two kits.

The quality and downstream functionality of the DNA and RNA obtained following protocols from the three kits was also assessed by comparing Ct values generated from a qPCR reaction. Figure 3 shows the average Ct values obtained at 10-fold dilution of the purified DNA and RNA when amplified using DNA- and RNA-specific primers for accurate assessment of each. In general, DNA and RNA extracted following Omega Bio-tek’s protocol amplified at lower cycle number suggesting better amplification potential and higher sensitivity compared to the nucleic acids extracted from Company T and Q. For instance, the 10-fold dilution of the purified DNA from the FFPE breast tumor tissue sample using Omega extraction amplified ~1.34 cycles lower than Company T’s and ~2.35 cycles lower than Company Q’s. Similarly, it was ~1.1 and ~1.16 cycles lower than Company T’s and ~0.9 and ~1.95 cycles lower than Company Q’s for FFPE breast and colorectal cancer tumor tissue samples, respectively (Figure 3).

The TapeStation analysis software enabled the assessment of DV200 values on the extracted RNA from the three FFPE tumor tissue samples and are as shown in Table 1. Irrespective of the extraction methodology, the DV200 value was greater than 20% satisfying Illumina’s requirement for RNA quality from an FFPE sample (Table 1). However, the results in Table 1 indicate that the RNA extracted using the Omega Bio-tek protocol has significantly higher percentage of fragments >200 nucleotides than the RNA extracted using Company T and Q’s kits indicating superior quality RNA (Breast – 74.9% with Omega vs 70.54% with Company T and 59.38 % with Company Q; Lung – 70.97% with Omega vs 66.75% with Company T and 38.4% with Company Q; Colorectal – 76.86% with Omega vs 69.85% with Company T and 60.28% with Company Q).
Table 1. Average DV200 value (percentage of fragments >200 nt) of RNA purified using different kits analyzed on Agilent’s TapeStation® 2200.
| FFPE Tumor Tissue Type | Kit Manufacturer | DV200 Region of Purified RNA |
|---|---|---|
| Breast | Omega Bio-tek | 74.90 |
| Company T | 70.54 | |
| Company Q | 59.38 | |
| Lung | Omega Bio-tek | 76.86 |
| Company T | 69.85 | |
| Company Q | 60.28 | |
| Colorectal | Omega Bio-tek | 70.97 |
| Company T | 66.75 | |
| Company Q | 38.40 |
Table 2 shows the ΔCq values estimated for FFPE colorectal tumor tissue samples relative to fresh frozen for the DNA purified using kits from Omega Bio-tek, Company T and Q. The ΔCq value with the Omega Bio-tek extraction was lower than those obtained using kits from Company T and Q. It is 3.10 with Omega compared to 4.06 and 5.32 with Company T and Company Q respectively. Lower ΔCq value with Omega Bio-tek indicates higher nucleic acid quality and truer representation of the actual tissue sample prior to FFPE processing. Illumina recommends a ΔCq <5 for optimal performance of TruSight Tumor 170 library [1] and the results indicate the DNA extracted from Omega Bio-tek and Company T satisfies this requirement whereas the purified DNA from Company Q fails this criterion.
Table 2. ΔCq values of DNA extracted from FFPE and non-FFPE colorectal tumo tissue samples using kits from different manufacturers.
| Kit Manufacturer | Colorectal Tissue Sample | Cq (or Ct) | ΔCq Relative to Fresh Frozen |
|---|---|---|---|
| Omega Bio-tek | FFPE | 25.42 | 3.10 |
| Company T | FFPE | 26.39 | 4.06 |
| Company Q | FFPE | 27.64 | 5.32 |
| N/A | Fresh Frozen | 22.33 | N/A |
For DNA extracted from different kits, we tracked the following sequencing metrics – median insert size (bp), % exon bases 250X coverage and % aligned reads (Table 3). All the extraction methodologies satisfy the quality threshold of 79bp for median insert size determined by Illumina for the TruSight™ 170 panel. The performance of the Omega Bio-tek extraction was equivalent to that of Company T’s and seems better in most metrics when compared to that of Company Q’s. For the colorectal tumor tissue sample, the quality metrics from Omega Bio-tek’s FFPE extraction were closer to that of the fresh frozen control. For instance, the median insert size of the control was 176 and Omega Bio-tek’s was 160 whereas it was 157 and 156 for Company T and Q respectively. The percentage of aligned reads and exon bases with at least 250X coverage follow similar suit. This suggests higher quality DNA extraction from FFPE samples using Omega Bio-tek’s chemistry and subsequently, a more accurate representation of the non-FFPE sample.
Figure 3. DNA from FFPE tumor samples was extracted, evaluated using the TruSight Tumor 170 assay, and sequenced on the HiSeq™ X System. The metrics of Median insert size, % Exon Bases 250X coverage and % aligned reads for different extraction methodologies are as listed below.
| FFPE Tumor Tissue Type | Kit Manufacturer | Median Insert Size | % Exon bases 250X | % Aligned Reads |
|---|---|---|---|---|
| Breast | Omega Bio-tek | 165 | 96.8 | 84.9 |
| Company T | 164 | 94.5 | 84.6 | |
| Company Q | 161 | 94.0 | 82.5 | |
| Lung | Omega Bio-tek | 162 | 93.9 | 83 |
| Company T | 162 | 89.4 | 83.5 | |
| Company Q | 149 | 79 | 53.8 | |
| Colorectal | Omega Bio-tek | 160 | 96.1 | 82 |
| Company T | 157 | 95.2 | 80.3 | |
| Company Q | 156 | 91.8 | 77.8 | |
| Fresh Frozen | 176 | 95.9 | 89.5 |
Figure 4 outlines the small variant calling analysis performed on two reported mutations in the TP53 and PIK3R1 genes in DNA extracted from FFPE colorectal tumor samples using the kits from Omega Bio-tek, Company T and Company Q and compared to the fresh frozen control. Figure 4 shows that the same mutations were called irrespective of the extraction kit used. However, comparing the allele frequencies and coverage between the fresh frozen control and FFPE sample for the mutations reported here, Omega extraction seems to be in most concordance with the control allowing for same conclusions to be drawn. Similar comparison results and conclusions were obtained post-sequencing on FFPE breast and lung tumor tissue samples (data not shown).

Conclusions
Omega Bio-tek’s Mag-Bind® FFPE DNA/RNA 96 Kit can extract both DNA and RNA from the same FFPE sample and exhibited superior performance when compared to comparable kits from Company T and Company Q both in terms of yield, quality as well as functionality. The results indicate better separation of DNA and RNA into separate eluates using Omega Bio-tek’s kit than the other two kits tested. The magnetic-bead-based approach of the Omega Bio-tek’s kit makes it automation and high-throughout friendly. This is a distinct advantage over Company Q’s kit which is column-based. The high quality of nucleic acids purified using Omega Bio-tek’s kit endorse their downstream functionality and corroborate their potential success in sequencing workflows.
References
1. Assessing DNA and RNA Quality from FFPE Samples for TruSight® Tumor 170. Illumina Technical Note – https://www.illumina.com/content/dam/illumina-marketing/documents/products/whitepapers/trusight-tumor-170-ffpe-white-pa-per-1170-2017-006.pdf
WP-0064
Related Products
-
Cultured Cells and Tissue
Mag-Bind® FFPE DNA/RNA 96 Kit
$0.00 – $2,443.40Price range: $0.00 through $2,443.40 Select options This product has multiple variants. The options may be chosen on the product page

