Carisa Townsend¹, Kiranmai Durvasula¹, Julie Baggs¹, and Travis Butts¹
¹Omega Bio-tek, Inc., Norcross, GA 30071
Introduction
Circulating free DNA (cfDNA) derived from urine has emerged as a promising biomarker in the field of non-invasive diagnostics, particularly in oncology, nephrology, and other areas of medical research. cfDNA is fragmented DNA present in the bloodstream or other body fluids, and its presence in urine offers several advantages over traditional biopsy and blood tests. One of the key benefits of cfDNA from urine is its non-invasive nature. Urinary cfDNA reflects both local and systemic genetic changes, offering insights into the underlying pathophysiology of diseases not only in the urinary tract but also in distant organs. This capacity to capture both localized and systemic alterations makes cfDNA an exciting tool for personalized medicine, where patient-specific molecular profiles can guide treatment decisions and improve outcomes. With advances in next-generation sequencing (NGS) and other molecular techniques such as droplet digital PCR (ddPCR), the analysis of cfDNA from urine holds the potential to revolutionize diagnostics, enabling earlier detection, better risk stratification, and more precise monitoring of therapeutic efficacy with minimal patient burden. Its integration into clinical practice could significantly enhance the management of various diseases, ultimately leading to improved patient outcomes.
To harness the full potential of cfDNA, important considerations should be allotted to 1) pre-analytical preservation to minimize cfDNA degradation post-sample collection, 2) reliable cfDNA extraction methodology to maximize cfDNA yield with minimal genomic DNA contamination and 3) appropriate downstream technology with high sensitivity to draw relevant conclusions. In this application note, we provide a complete workflow solution from urine sample collection and preservation to cfDNA extraction and subsequent implementation in droplet digital PCR downstream for detection of rare variant alleles.
Colli-Pee™* UAS™ preservative from DNA Genotek Inc. was used to preserve the urine sample and cfDNA was extracted using Omega Bio-tek’s Mag-Bind® cfDNA Kit (M3298). Here, we present the workings of this workflow for extraction of cfDNA from 4 mL input volume with supporting cfDNA yield information and further demonstrate the use of extracted cfDNA in ddPCR application for detection of NRAS and EGFR gene variants at different allelic frequencies.
*For Research Use Only. Not for Diagnostic Use.
Learn more about the Mag-Bind® cfDNA Kit.

Materials and Methods
Sample Collection and Extraction
Urine sample from an anonymous female donor was collected and added to DNA Genotek’s Colli-Pee™ UAS™ preservative at a ratio 2.3 to 1 for sample preservation. To mimic a clinical sample, Mimix™ Multiplex I cfDNA Set Reference Standard (HD780) obtained from Horizon Discovery was spiked into the preserved urine sample at different allelic frequencies. Horizon Discovery’s reference standard consists of 4 vials containing 8 onco-relevant mutations at different allelic frequencies, 5%, 1%, 0.1% and 0% (100% Wildtype (WT)). 100 µL (350 ng) from each of the vials of the reference standard standards were independently spiked into the preserved urine sample to measure extraction efficiency and assay performance. The experimental groups included in this study included 1) Reference standard at different allelic frequencies spiked into UAS preserved urine, 2) UAS preserved urine (No spike-in control), 3) Reference standard at different allelic frequencies spiked into UAS preservative (No urine control) 4) Reference standard at different allelic frequencies spiked into elution buffer (No urine, no preservative control). All the samples were centrifuged at 4,000 g for 15 minutes before the start of the protocol. Omega Bio-tek’s Mag-Bind® cfDNA Kit following a modified protocol for urine samples was used for the extraction of cfDNA from 4 mL input volume from each of the four experimental groups described above and eluted in 50 µL volume.
Yield and Quality
The yield and quality of the cfDNA was evaluated using the Cell-Free DNA ScreenTape Assay on the Agilent 4150 TapeStation system and its fraction relative to the total DNA extracted was assessed.
Droplet Digital PCR
cfDNA purified from 4 mL inputs containing oncogenes at 4 allelic frequencies (5%, 1%, 0.1% and 0%) was subjected to droplet digital PCR on Bio-Rad QX200 Droplet Digital PCR System. Two assays were developed by detecting NRAS (variant: A59T) and EGFR (variant: L858R) genes using ddPCR utilizing FAM probe for the mutant allele and SUN probe for the wildtype. 15 ng of cfDNA was mixed with Bio-Rad’s ddPCR Supermix for Probes (No dUTP) (Bio-Rad, Hercules, CA, USA) in a total reaction volume of 300 µL according to the manufacturer’s protocol. The droplets were generated using Bio-Rad’s QX200 Droplet Generator following manufacturer’s instruction and subjected to amplification on the C1000 Touch Thermal Cycler with adjustments of annealing temperature for each probe. The droplets were scanned using QX200 QuantaSoft Droplet Reader (Bio-Rad) and data was analyzed using Bio-Rad’s QuantaSoft Analysis Pro Software. ddPCR work and data analysis were performed externally at Emory University, Atlanta, GA.
Results and Discussion
TapeStation Analysis of Purified cfDNA
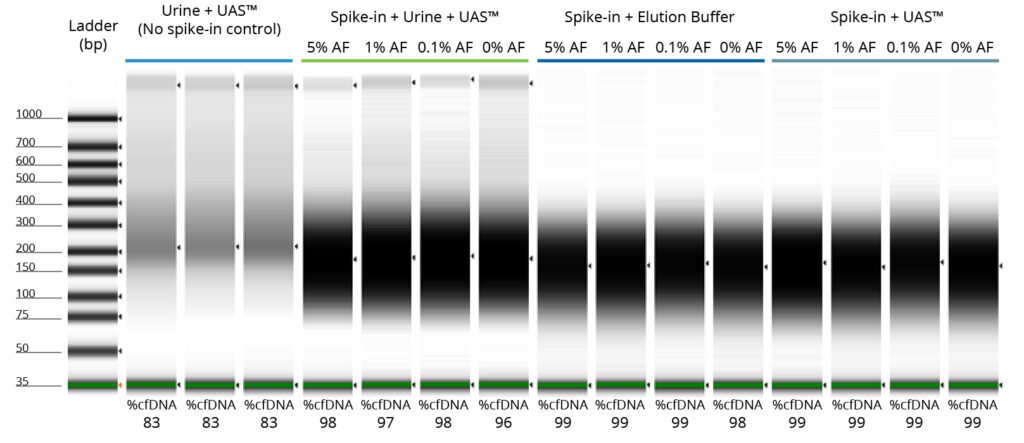
TapeStation analysis using the Cell-Free DNA ScreenTape Assay was performed to evaluate the yield and quality of extracted cfDNA from all the experimental groups as described above. Dark bands concentrated between 150 and 200 base pairs indicate high yielding, high quality cfDNA extracted from each sample and all the experimental groups tested (Figure 1). In Figure 1, the samples labelled 0% AF represents the 100% wildtype sample which is the control, while the remaining samples are labelled according to the allelic frequency (5%, 1%, and 0.1%) of the reference standard used. As expected, there is endogenous cfDNA from urine present in the no spike-in control with just the preserved urine in Colli-Pee™ UAS™ Preservative. There is a presence of high molecular weight band indicative of genomic DNA in this experimental group and the estimated cfDNA in the extract was 83%. This higher molecular weight gDNA band was also present in preserved urine samples with spike-ins at different allelic frequencies, suggestions that the extraction kit likely extracted the endogenous cfDNA and gDNA present in
Table 1. cfDNA Yield as estimated by TapeStation Analysis.
| Experimental Group | Average Estimated Yield (ng) |
|---|---|
|
Urine + UAS™ (No spike-in control) |
58.2 |
| Spike-in + Urine + UAS™ | 226.5 |
|
Spike-in + Elution Buffer (No urine, no preservative control) |
161.9 |
|
Spike-in + UAS™ (No urine control) |
186.1 |
urine along with externally spiked-in cfDNA. No urine/no preservative and no urine controls resulted in high-yielding cfDNA just from the spike-ins and predictably the higher molecular weight band is absent (i.e. no genomic DNA) since there is no actual urine sample.
Table 1 shows the average cfDNA yield as estimated by the TapeStation® analysis software within the 100-700 bp region for the Cell-Free DNA ScreenTape Assay for all the four experimental groups tested. The endogenous cfDNA yield in the urine samples preserved in Colli-Pee™ UAS™ Preservative was 58.2 ng. The experimental groups with the spike-in’s in either elution buffer or Colli-Pee™ UAS™ Preservative were ~161.9 ng or 186.1 ng respectively, indicating a % recovery of 46-53% based on a spike-in input of 350 ng to each sample. For the experimental condition with urine preserved in Colli-Pee™ UAS™ Preservative spiked-in with reference standards, the average yield was 226.5 ng attesting to the extraction of both endogenous cfDNA as well as the spiked-in cfDNA.
NRAS Droplet Populations

UAS: Colli-pee™ UAS™ preservative
EGFR Droplet Populations
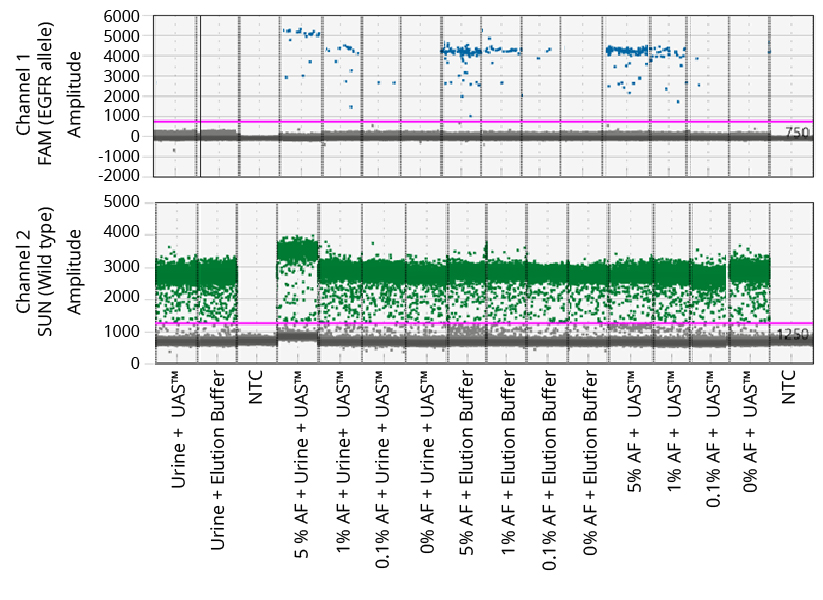
UAS: Colli-pee™ UAS™ preservative
Concordance Analysis of Observed vs Expected Allelic Frequencies for NRAS Assay
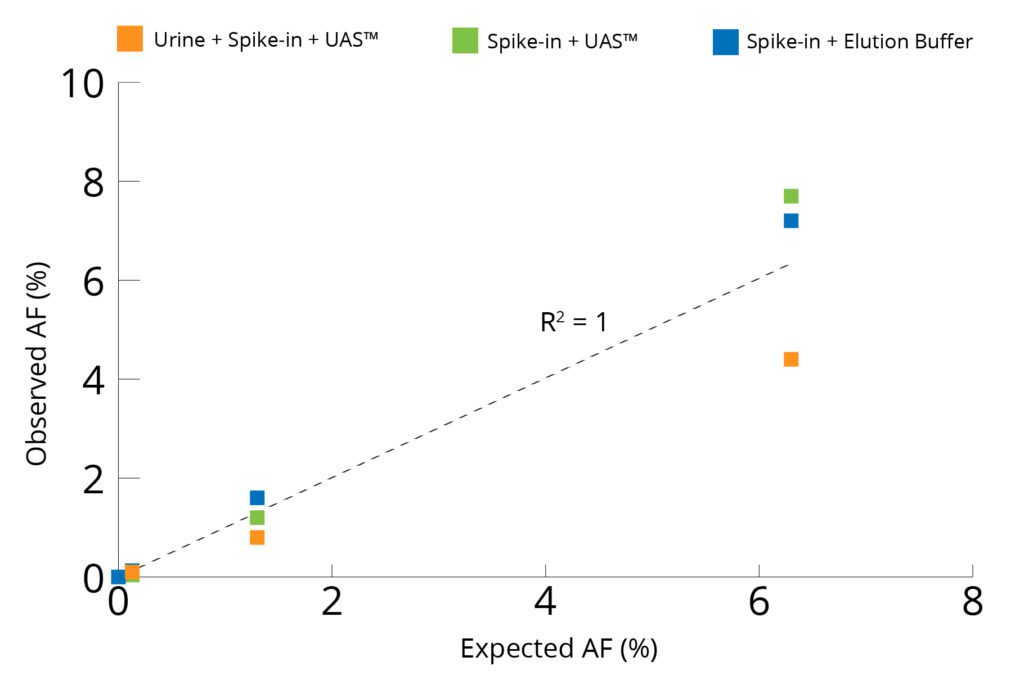
AF: Allelic Frequency. UAS: Colli-pee™ UAS™ preservative
Concordance Analysis of Observed vs Expected Allelic Frequencies for EGFR Assay

AF: Allelic Frequency. UAS: Colli-pee™ UAS™ preservative
The two ddPCR assays (NRAS and EGFR) performed on the cfDNA extracted from each of the four experimental groups generated approximately 15,000 droplets. There was no significant difference in the number of droplets generated from urine samples preserved with Colli-Pee™ UAS preservative compared to elution buffer alone. The droplet populations at each allelic frequency in the NRAS and EGFR assays are shown in Figures 2 & 3, respectively. The threshold for each assay was manually set based on the droplet population of 0% AF (100% wildtype) control making sure there is a clear separation between the positive and negative populations. The absence of droplets in the No Template Control (NTC) confirms no reagent contamination. The absence of positive droplets in the No Spike-in control group indicates no non-specific amplification. The assay results for NRAS and EGFR genes at each allelic frequency were then interpreted based on the threshold set. Positive droplets were detected for both NRAS and EGFR assays at all the allelic frequencies tested.
The number of positive droplets at each allelic frequency was summed and compared to the expected allelic frequency to check for concordance between actual vs expected. The results of the concordance analysis for NRAS and EGFR genes are shown in Figures 4 and 5, respectively. The observed allelic frequencies for NRAS gene are in concordance with the expected allelic frequencies for all the experimental groups tested and well within the acceptance criteria mentioned on the Certificate of Analysis of the reference standards (spike-in) set by the manufacturer. A similar trend was observed with EGFR assay except for when reference standard at 5% allelic frequency was spiked into urine preserved in Colli-Pee™ UAS™ Preservative. The observed allelic frequency was lower at 2.5% where the acceptance criteria was between 3.5 and 6.5%, likely an experimental error. For all the other experimental groups, EGFR assay shows excellent concordance between observed and expected values. The results indicate cfDNA preserved in Colli-Pee™ UAS™ Preservative and extracted using Mag-Bind® cfDNA Kit (M3298) was capable of detection of mutant alleles at frequencies as low as 0.1% when used in ddPCR workflow.
Conclusions
The complete workflow from preservation with Colli-Pee™ UAS™ preservative to extraction of cfDNA from preserved urine using Omega Bio-tek’s Mag-Bind® cfDNA Kit (M3298) resulted in high-quality cfDNA suitable for use in downstream applications like ddPCR and was able to detect mutant alleles at frequencies as low as 0.1%. The combination of urine sample preservation, high sensitivity cfDNA purification, and ability to detect low-frequency mutations using ddPCR makes this an attractive solution for cfDNA analysis in clinical and research settings.
Disclaimer
This study was supported in part by the Emory Integrated Genomics Core (EIGC) (RRID:SCR_023529), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
WP-0061
Related Products
-
Circulating DNA
Mag-Bind® cfDNA Kit
$0.00 – $2,675.20Price range: $0.00 through $2,675.20 Select options This product has multiple variants. The options may be chosen on the product page



