Carisa Townsend1, Kiranmai Durvasula1, Julie Baggs1, and Travis Butts1
- Omega Bio-tek, Inc, Norcross, GA 30071
Introduction
Recent advancements and studies have demonstrated the potential of cfDNA as a universal, non-invasive biomarker in cancer prognosis, diagnosis, and prenatal testing. cfDNA are found in low abundance and in a background of contaminating genomic DNA making it a challenging testing analyte. To harness the full potential of cfDNA, important considerations should be allotted to reliable cfDNA extraction methodology to maximize cfDNA yield with minimal genomic DNA contamination and appropriate downstream technology with high sensitivity to draw relevant conclusions. In this application note, we aim to equip researchers with a simple and efficient semi-automated cfDNA extraction workflow and present droplet digital PCR (ddPCR) as a powerful tool for detecting ultra-low variants with high specificity.
cfDNA extraction was achieved by employing Omega Bio-tek’s Mag-Bind® cfDNA Kit (M3298) automated on MagBinder® Fit24 Nucleic Acid Purification System. The MagBinder® Fit24 is a new nucleic acid purification system designed to meet the needs of low-to-moderate throughput users by providing semi-automated purification of 1 to 24 small or large volume sample inputs. Here, we present the workings of this workflow for extraction of cfDNA from 4 mL working volume with supporting cfDNA yield information and further demonstrate the use of extracted cfDNA in ddPCR application for detection of NRAS and EGFR genes at different allelic frequencies.
Materials and Methods
Sample
Multiplex I cfDNA in Synthetic Matrix II (HD917) obtained from Horizon Discovery was used to mimic a clinical sample. 4 vials containing 8 onco-relevant mutations at different allelic frequencies, 5%, 1%, 0.1% and 0% (100% Wildtype (WT)), were provided with the reference set to measure extraction efficiency and assay performance. 100% WT from the reference set would serve as a control.
Extraction Kit
Omega Bio-tek’s Mag-Bind® cfDNA Kit was used for the extraction of cfDNA from 4 mL input volume. Briefly, 1 mL from each of the 4 vials of Multiplex I cfDNA in Synthetic Matrix was combined with 3 mL of elution buffer included with the kit to bring the starting sample volume to 4 mL. The reagents from the Mag-Bind® cfDNA Kit were user-filled into a specially designed MB Fit24™ cartridge (MB Fit24™ Reagent cartridge, 10 mL) in volumes and positions as shown in Figure 1. Omega Bio-tek also provides a prefilled option in MB Fit24™ cfDNA Kit (B3298-10-48PF) for cfDNA extraction using MagBinder® Fit24 with reagents pre-arrayed in cartridges for ease-of-use and minimal manual intervention.
Contents, Volumes, and Positions of Dispensed Reagents
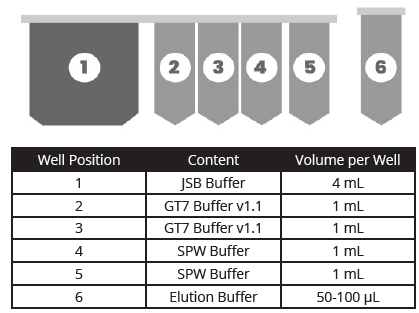
Figure 1. MagBinder® Fit24 reagent cartridge positions, contents, and volumes.
Extraction Methodology
The user-filled reagent cartridges were loaded on the MagBinder® Fit24 and the preloaded script for Mag-Bind® cfDNA Kit was selected and ran on the instrument. All samples were run on the MagBinder® Fit24 platform simultaneously with the magnetic rods working in unison to pick-up, transfer and release magnetic particles in different wells of the reagent cartridge to provide purified cfDNA in the elution tube at the end. cfDNA was eluted in 100 μL volume and the total protocol time on the MagBinder® Fit24 platform was ~54 minutes from cartridge placement to cfDNA elution.
Yield and Quality
The yield and quality of the cfDNA was evaluated using the Cell-Free DNA ScreenTape Assay on the Agilent 4150 TapeStation® system and its fraction relative to the total DNA extracted was assessed.
Droplet Digital PCR
cfDNA purified from 4 mL inputs containing oncogenes at 4 allelic frequencies (5%, 1%, 0.1% and 0%) was subjected to droplet digital PCR on Bio-Rad QX200 Droplet Digital PCR System. Two assays were developed by detecting NRAS and EGFR mutations using ddPCR utilizing FAM probe for the mutant allele and SUN probe for the wild type. 15 ng of cfDNA was mixed with Bio-Rad’s ddPCR Supermix for Probes (No dUTP) (Bio-Rad, Hercules, CA, USA) in a total reaction volume of 300 μL according to the manufacturer’s protocol. The droplets were generated using Bio-Rad’s QX200 Droplet Generator following manufacturer’s instruction and subjected to amplification on the C1000 Touch Thermal Cycler with adjustments of annealing temperature for each probe. The droplets were scanned using QX200 QuantaSoft Droplet Reader (Bio-Rad) and data was analyzed using Bio-Rad’s QuantaSoft Analysis Pro Software. ddPCR work and data analysis was performed externally at Emory University, Atlanta, GA.
Results and Discussion
TapeStation analysis using the Cell-Free DNA ScreenTape Assay was performed to evaluate the yield and quality of extracted cfDNA from sample inputs containing 4 different allelic frequencies. Dark bands concentrated between 150 and 200 base pairs indicate high yielding, high-quality extracted cfDNA for each sample (Figure 2). Here, the sample labelled 100% WT represents the 100% wild type sample which is the control, while the remaining samples are labelled according to the allelic frequency (5%, 1%, and 0.1%) of the reference standard used. The cfDNA yield as calculated by the TapeStation analysis software within the 50-700bp region for the Cell-Free DNA ScreenTape Assay for all four extractions ranged between 238.4 ng to 285.6 ng which is significantly greater than the Horizon Discovery’s reported expected yield of 120 ng per 1 mL of product using the QIAmp® Circulating Nucleic Acid Kit with 2 mL extraction input1. From their certificate of analysis, Horizon Discovery expects 30% extraction efficiency; whereas the MagBinder® Fit24 extraction workflow was able to achieve 60-70% extraction efficiency.
TapeStation Analysis of Purified cfDNA

Allele Populations in NRAS Assay
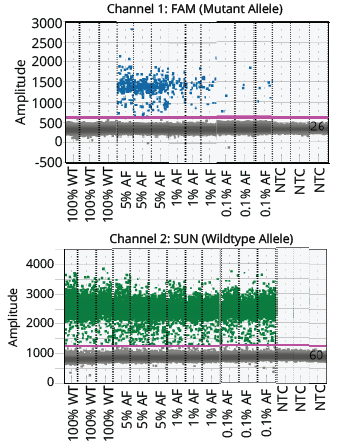
Figure 3. Visualization of the positive and negative populations of the NRAS assay. The threshold is indicated in the graph with a pink horizontal line.
Allele Populations in EGFR Assay
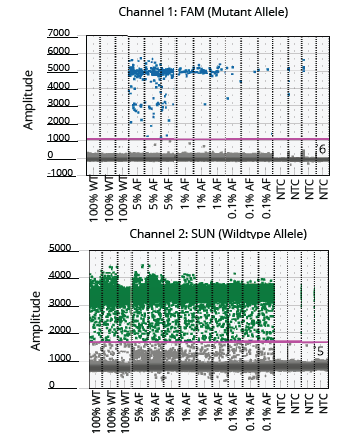
Figure 5. Visualization of the positive and negative droplet populations of the EGFR assay. The threshold is indicated with a pink horizontal line.
NRAS allele Ratio Determined by ddPCR vs Expected
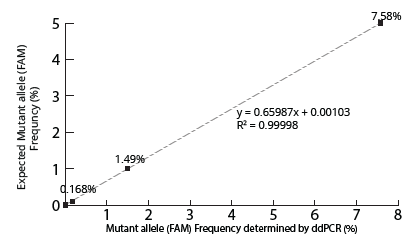
Figure 4. Concordance analysis of NRAS gene at allelic frequencies determined by ddPCR vs expected. NRAS mutant allele ratio presents as expected.
EGFR Allele Ratio Determined by ddPCR vs Expected

Figure 6. Concordance analysis of EGFR gene at allelic frequencies determined by ddPCR vs expected. EGFR mutant allele ratio presents as expected.
The two ddPCR assays (NRAS and EGFR) performed on the cfDNA extracted from each of the four samples generated approximately 50,000 droplets. The droplet populations at each allelic frequency in the NRAS and EGFR assays are shown in Figures 3 & 5, respectively. The threshold for each assay was manually set based on the droplet population of 100% Wildtype control making sure there is a clear separation between the positive and negative populations. The absence of droplets in the No Template Control (NTC) confirms no non-specific amplification. The assay results for NRAS and EGFR genes at each allelic frequency was then interpreted based on the threshold set. Positive droplets were detected for both NRAS and EGFR assays at all the allelic frequencies tested. The number of positive droplets at each allelic frequency was summed and compared to the expected allelic frequency to check for concordance between actual vs expected. The results of the concordance analysis for NRAS and EGFR genes are shown in Figures 4 & 6 respectively. The expected ratios are 0 for 100% Wildtype and 5%, 1%, and 0.1% for the mutant alleles. The 5%, 1% and 0.1% samples have an experimental ratio of approximately 7.6%, 1.5% and 0.17% in the NRAS assay (Figure 4) and 4.66%, 1.12%, and 0.17% samples in the EGFR assay (Figure 6) respectively. These results demonstrate ddPCR assay was successful and shows excellent concordance between the actual and expected values. The results indicate cfDNA extracted using Mag-Bind® cfDNA Kit (M3298) automated on MagBinder® Fit24 Nucleic Acid Purification System when used in ddPCR workflow was capable of detection of mutant alleles at frequencies as low as 0.1%.
Conclusions
ddPCR is a robust method for quantitative analysis of multiple target genes within one sample that is best exploited when an efficient and reliable workflow for nucleic acid extraction is in place. The semi-automated workflow for extraction of cfDNA using Mag-Bind® cfDNA Kit (M3298) on MagBinder® Fit24 Nucleic Acid Purification System is capable of isolating high yields of high-quality cfDNA, as exhibited by the kit’s high extraction efficiency of 60-70%. In addition to this, the detection of mutant alleles at frequencies as low as 0.1% from cfDNA extracted using this workflow confirm the Mag-Bind® cfDNA Kit and the MagBinder® Fit24‘s capability of extracting cfDNA suitable for use in downstream applications such as ddPCR.
Disclaimer
This study was supported in part by the Emory Integrated Genomics Core (EIGC) (RRID:SCR_023529), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
References
1. Horizon Discovery. Multiplex cfDNA in Synthetic Matrix II Reference Standard: Certificate of Analysis. URL: https://horizondiscovery.com/-/media/Files/Horizon/resources/Certificate-of-analysis/HD917_45462_V2.pdf?sc_lang=en
Related Products
-
Circulating DNA
Mag-Bind® cfDNA Kit
$0.00 – $2,675.20Price range: $0.00 through $2,675.20 Select options This product has multiple variants. The options may be chosen on the product page